InnoVar simulations
Danilo Sarti
2022-11-24
innovar_flow.RmdLoading Packages
library(InnoVaR)
#> Warning: replacing previous import 'jmuOutlier::rlaplace' by 'rmutil::rlaplace'
#> when loading 'InnoVaR'
#> Warning: replacing previous import 'jmuOutlier::qlaplace' by 'rmutil::qlaplace'
#> when loading 'InnoVaR'
#> Warning: replacing previous import 'jmuOutlier::plaplace' by 'rmutil::plaplace'
#> when loading 'InnoVaR'
#> Warning: replacing previous import 'jmuOutlier::dlaplace' by 'rmutil::dlaplace'
#> when loading 'InnoVaR'
#> Warning: replacing previous import 'copula::profile' by 'stats::profile' when
#> loading 'InnoVaR'
#> Warning: replacing previous import 'Matrix::cov2cor' by 'stats::cov2cor' when
#> loading 'InnoVaR'
#> Warning: replacing previous import 'rmutil::nobs' by 'stats::nobs' when loading
#> 'InnoVaR'
#> Warning: replacing previous import 'dplyr::filter' by 'stats::filter' when
#> loading 'InnoVaR'
#> Warning: replacing previous import 'dplyr::lag' by 'stats::lag' when loading
#> 'InnoVaR'
#> Warning: replacing previous import 'Matrix::toeplitz' by 'stats::toeplitz' when
#> loading 'InnoVaR'
#> Warning: replacing previous import 'copula::coef' by 'stats::coef' when loading
#> 'InnoVaR'
#> Warning: replacing previous import 'Matrix::update' by 'stats::update' when
#> loading 'InnoVaR'
#> Warning: replacing previous import 'copula::logLik' by 'stats::logLik' when
#> loading 'InnoVaR'
#> Warning: replacing previous import 'copula::confint' by 'stats::confint' when
#> loading 'InnoVaR'
library(tidyverse)
library(ggplot2)
library(readxl)
library(readr)
library(copula)
library(mice)
library(checkmate)
library(jmuOutlier)
library(ggpubr)Loading datasets containing the attributes of the modules to be simulated
load("/Volumes/T7 Touch/InnoVaR/data/gen_att.rda")
head(gen_att)
#> # A tibble: 6 × 100
#> trait_1 trait_2 trait_3 trait_4 trait_5 trait_6 trait_7 trait_8 trait_9
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 0 0 0 0 0 0 0 0 0
#> 2 1 1 1 1 1 1 1 1 1
#> 3 2 2 2 2 2 2 2 2 2
#> 4 NA NA NA NA NA NA NA NA NA
#> 5 NA NA NA NA NA NA NA NA NA
#> 6 NA NA NA NA NA NA NA NA NA
#> # … with 91 more variables: trait_10 <chr>, trait_11 <chr>, trait_12 <chr>,
#> # trait_13 <chr>, trait_14 <chr>, trait_15 <chr>, trait_16 <chr>,
#> # trait_17 <chr>, trait_18 <chr>, trait_19 <chr>, trait_20 <chr>,
#> # trait_21 <chr>, trait_22 <chr>, trait_23 <chr>, trait_24 <chr>,
#> # trait_25 <chr>, trait_26 <chr>, trait_27 <chr>, trait_28 <chr>,
#> # trait_29 <chr>, trait_30 <chr>, trait_31 <chr>, trait_32 <chr>,
#> # trait_33 <chr>, trait_34 <chr>, trait_35 <chr>, trait_36 <chr>, …
load("/Volumes/T7 Touch/InnoVaR/data/soil_att_innovar.rda")
head(soil_att_innovar)
#> # A tibble: 6 × 129
#> yes_n…¹ siteid Drana…² Weath…³ Weath…⁴ USDAT…⁵ USDAM…⁶ Physi…⁷ Major…⁸ Major…⁹
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 Yes 1 Very p… Sunny/… No rai… Pergel… Aquic Crest … Level … Plain
#> 2 No 2 Poorly… Partly… No rai… Cryic Udic Upper … Slopin… Plateau
#> 3 NA 3 Imperf… Overca… No rai… Frigid Ustic Middle… Steep … Depres…
#> 4 NA 4 Modera… Rain Rainy … Mesic Xeric Lower … NA Valley…
#> 5 NA 5 Well d… Sleet Heavie… Thermic Aridic Toe sl… NA Medium…
#> 6 NA 6 Somewh… Snow Extrem… Hypert… Peraqu… Bottom… NA Medium…
#> # … with 119 more variables: LandformComplexValue <chr>,
#> # SlopePathwaysValue <chr>, SlopeFormValue <chr>, SlopeGradientValue <chr>,
#> # SlopeOrientation <chr>, LandUseClassValue1 <chr>, LandUseClassValue2 <chr>,
#> # LandUseClassValue3 <chr>, CropClassValue1 <chr>, CropClassValue2 <chr>,
#> # HumanInfluenceClassValue <chr>, VegetationClassValue1 <chr>,
#> # VegetationClassValue2 <chr>, LithologyValueClass <chr>,
#> # LithologyValueGroup <chr>, LithologyValueType <chr>, …
soil_att_ex=soil_att_innovarSimulating soil module:
declaring global information
## declaring the number of variables in the module
n_var_soil=ncol(soil_att_ex)
## filtering the categorical ones.
soil_cat=soil_att_ex %>% select(which(sapply(soil_att_ex, class) == 'character'))
## filtering the numerical values.
soil_num=soil_att_ex %>% select(which(sapply(soil_att_ex, class) != 'character'))Informing correlations known a priori
known_neg_dis=calculate_trig_dis(c(-0.8016201, -0.8022176, -0.7633010))
known_pos_dis=calculate_trig_dis(c(0.5,0.6,0.9))creating a data frame to store
n_var=ncol(soil_att_ex)
soil_coordinates=module_coords(n_var=ncol(soil_att_ex),
which_neg=c("2_1","6_2","1_9","1_6","2_3"),
dis_neg_know_ass=known_neg_dis,
coo_known_neg_ass=c("2_1","6_2","1_9"),
dis_pos_know_ass=known_pos_dis,
coo_known_pos_ass=c("3_9","4_5","6_8"),
by=0.00001## this should be small
)Assuring correlation matrices
soil_corr=round(assure_corr(n_var=n_var,corr=sym_sin_soil))
library(matrixcalc)
soil_corr=as.matrix(soil_corr)
#Checking if its positive definite.. if you round the matrix it can be not positie definite.
is.positive.definite(soil_corr)
#> [1] TRUESetting a copula to simulation
sim_data_cop=set_sim_copula(d=n_var,
lower_tri_corr=soil_corr[lower.tri(soil_corr,diag=FALSE)],
n_cont_var=ncol(soil_num),
cont_var_par=list(list(lambda=40),list(df=30,ncp=3),
list(shape=40,scale=10),list(rate=30),list(lambda=40),list(df=30,ncp=3),
list(shape=40,scale=10),list(rate=30),list(lambda=40),list(df=30,ncp=3),
list(shape=40,scale=10),list(rate=30),list(lambda=40),list(df=30,ncp=3),
list(shape=40,scale=10)) ,
n_unique=length(unique(na.omit(soil_att_ex$siteid))),
mar_cont_dists=c("pois","chisq","gamma","exp","pois","chisq","gamma","exp","pois","chisq","gamma","exp","pois","chisq","gamma"),
var_names=colnames(soil_att_ex)
)
###
cut_data_soil=cut_levels(categorical=soil_cat,
cat_inc_all_levels=c("siteid"),
levels_inc_all=list(siteid=c(1:16)), continuous_to_cut=sim_data_cop$simulated
)
head(cut_data_soil)
#> yes_no_sample DranaigeClass WeatherConditionsValue_present
#> 1 Yes Somewhat excessively drained Sleet
#> 2 Yes Well drained Rain
#> 3 No Poorly drained Overcast
#> 4 No Imperfectly drained Sleet
#> 5 Yes Somewhat excessively drained Snow
#> 6 Yes Excessively drained Sunny/clear
#> WeatherConditionsValue_former USDATempCode USDAMoistureCode
#> 1 Extremely rainy time or snow melting Isomesic Ustic
#> 2 No rain in the last month Hyperthermic Xeric
#> 3 Extremely rainy time or snow melting Isofrigid Perudic
#> 4 Rainy without heavy rain in the last 24 hours Pergelic Peraquic
#> 5 Rainy without heavy rain in the last 24 hours Pergelic Aquic
#> 6 No rain in the last week Isofrigid Aridic
#> PhysiographyValue MajorLandformValue1 MajorLandformValue2
#> 1 Intermediate part (talf) Steep land Medium-gradient hill
#> 2 Bottom (drainage line) Sloping land High-gradient valley
#> 3 Lower slope (foot slope) Level land Medium-gradient escarpment zone
#> 4 Higher part (rise) Level land Medium-gradient escarpment zone
#> 5 Lower part (and dip) Level land Plateau
#> 6 Lower part (and dip) Sloping land High-gradient hill
#> LandformComplexValue SlopePathwaysValue SlopeFormValue
#> 1 Dome-shaped SV Complex (Irregular)
#> 2 Ridged SS Terraced
#> 3 Cuesta-shaped SS Complex (Irregular)
#> 4 Terraced SS Terraced
#> 5 Dome-shaped VS Complex (Irregular)
#> 6 Strong karst VS Straight
#> SlopeGradientValue SlopeOrientation LandUseClassValue1
#> 1 Very gently sloping South Not used and not managed
#> 2 Sloping South-west Forestry
#> 3 Gently sloping South-south-west Mixed farming
#> 4 Very gently sloping East-north-east Crop agriculture (cropping)
#> 5 Sloping North-north-west Nature protection
#> 6 Very gently sloping South-west Not used and not managed
#> LandUseClassValue2 LandUseClassValue3 CropClassValue1
#> 1 Nature and game preservation Animal production Oilcrops
#> 2 Degradation control Reserves Fruits and melons
#> 3 Annual field cropping Clear felling Fodder plants
#> 4 Transport Shifting cultivation Oilcrops
#> 5 Nature and game preservation Ranching Fodder plants
#> 6 Nature and game preservation With interference Oilcrops
#> CropClassValue2 HumanInfluenceClassValue VegetationClassValue1
#> 1 Coffee No influence Closed forest
#> 2 Sugar cane Surface compaction Woodland
#> 3 Leguminous Drip irrigation Rainwater-fed moor peat
#> 4 Melons Sand additions Woodland
#> 5 Maize (fodder plant) Artificial drainage Herbaceous
#> 6 Apples Burning Groundwater-fed bog peat
#> VegetationClassValue2 LithologyValueClass
#> 1 Forb Sedimentary rock (consolidated)
#> 2 Evergreen shrub Sedimentary rock (unconsolidated)
#> 3 Evergreen broad-leaved forest Sedimentary rock (consolidated)
#> 4 Deciduous forest Igneous rock
#> 5 Xeromorphic shrub Igneous rock
#> 6 Semi-deciduous dwarf shrub Sedimentary rock (unconsolidated)
#> LithologyValueGroup LithologyValueType SurfaceAgeValue RockOutcropsCoverValue
#> 1 UC UC1 Ya Dominant
#> 2 UU MA3 O Many
#> 3 UU MB3 O Very few
#> 4 UE SE1 Yn None
#> 5 UE UF2 O Common
#> 6 UE IA1 vYa Very few
#> RockOutcropsDistanceValue FragmentCoverValue FragmentsSizeValue
#> 1 20–50 m None Boulders
#> 2 2–5 m Abundant Large boulders
#> 3 5–20 m Common Large boulders
#> 4 20–50 m None Medium gravel
#> 5 < 2 m Abundant Stones
#> 6 > 50 m Dominant Stones
#> ErosionCategoryValue1 ErosionCategoryValue2
#> 1 Wind and water erosion Sheet erosion
#> 2 Water erosion or deposition Wind deposition
#> 3 Wind and water erosion Shifting sands
#> 4 Not known Rill erosion
#> 5 Wind and water erosion Salt deposition
#> 6 Wind (aeolian) erosion or deposition Wind deposition
#> ErosionTotalAreaAffectedValue ErosionDegreeValue ErosionActivityPeriodValue
#> 1 25–50 % Severe Active in historical times
#> 2 10–25 % Severe Active in recent past
#> 3 25–50 % Severe Period of activity not known
#> 4 0–5 % Moderate Active in recent past
#> 5 5–10 % Severe Period of activity not known
#> 6 10–25 % Extreme Active in historical times
#> SealingThicknessValue SealingConsistenceValue CracksWidthValue
#> 1 Medium Slightly hard Medium (1 - 2 cm)
#> 2 Medium Hard Medium (1 - 2 cm)
#> 3 Very thick Slightly hard Fine (< 1 cm)
#> 4 None Slightly hard Very wide (5 - 10 cm)
#> 5 Very thick Extremely hard Extremely wide (>10 cm)
#> 6 None Hard Very wide (5 - 10 cm)
#> CracksDepthValue CracksDistanceValue SaltCoverValue
#> 1 Surface (< 2 cm) Widely spaced (2 - 5 m) None (0 - 2 %)
#> 2 Medium (2 - 10 cm) Widely spaced (2 - 5 m) High (40 - 80 %)
#> 3 Very deep (> 20 cm) Very closely spaced (< 0,2 m) High (40 - 80 %)
#> 4 Very deep (> 20 cm) Very closely spaced (< 0,2 m) High (40 - 80 %)
#> 5 Deep (10 - 20 cm) Closely space (0,2 - 0,5 m) Dominant (> 80%)
#> 6 Medium (2 - 10 cm) Very closely spaced (< 0,2 m) High (40 - 80 %)
#> SaltThicknessValue BleachedSandValue BoundaryDistinctnessValue
#> 1 Very thick (> 20 mm) Dominant (> 80%) Abrupt (0 - 2 cm)
#> 2 Very thick (> 20 mm) None (0 - 2 %) Diffuse (> 15 cm)
#> 3 None (-) Dominant (> 80%) Abrupt (0 - 2 cm)
#> 4 Thick (5 -20 mm) Moderate (15 - 40 %) Clear (2 - 5 cm)
#> 5 Medium (2 - 5 mm) Low (2 - 15 %) Gradual (5 - 15 cm)
#> 6 Thick (5 -20 mm) Low (2 - 15 %) Gradual (5 - 15 cm)
#> BoundaryTopographyValue FieldTexture TextureValue
#> 1 Irregular; pockets more deep than wide Sand Sandy loam
#> 2 Irregular; pockets more deep than wide Silt Sandy clay
#> 3 Irregular; pockets more deep than wide Clay Loamy sand
#> 4 Smooth; nearly plane surface Silt Clay
#> 5 Irregular; pockets more deep than wide Clay Sandy loam
#> 6 Wavy; pockets less deep than wide Silt Silt loam
#> SandyTextureValue TextureClassValue ArtefactsAbundance
#> 1 Sand, unsorted Sandy clay loam Stone line
#> 2 Very fine sand Silty clay Few (2 - 5 %)
#> 3 Sand, unsorted Sandy clay loam Many (15 - 40%)
#> 4 Fine sand Very coarse and coarse sand Common (5 -15 %)
#> 5 Sand, unsorted Heavy clay None (0 %)
#> 6 Medium sand Silty clay loam Common (5 -15 %)
#> RockSizeValue2 ArtefactsSizeValue
#> 1 Fine gravel (2 - 6 mm) Medium artefacts (6 - 20 mm)
#> 2 Large boulders (> 600 mm) Coarse artefacts (> 20 mm)
#> 3 Large boulders (> 600 mm) Medium artefacts (6 - 20 mm)
#> 4 Large boulders (> 600 mm) Medium artefacts (6 - 20 mm)
#> 5 Stones (60 - 200 mm) Fine artefacts (2 - 6 mm)
#> 6 Stones (60 - 200 mm) Fine artefacts (2 - 6 mm)
#> ArtefactsSizeCombi RockShapeValue ArtefactsWeatheringValue
#> 1 Fine and medium gravel/artefacts Angular Fresh or slightly weathered
#> 2 Medium and coarse gravel/artefacts Rounded Fresh or slightly weathered
#> 3 Medium and coarse gravel/artefacts Flat Weathered
#> 4 Fine and medium gravel/artefacts Angular Strongly weathered
#> 5 Coarse gravel and stones Flat Weathered
#> 6 Boulders and large boulders Flat Fresh or slightly weathered
#> MineralFragmentsValue PeatDecompostionValue AbundanceValue
#> 1 Mica D2 Very few (0 - 2 %)
#> 2 Feldspar D2 Common (5 -15 %)
#> 3 Mica D3 Very few (0 - 2 %)
#> 4 Quartz D1 None (-)
#> 5 Feldspar D5.1 Very few (0 - 2 %)
#> 6 Feldspar D2 Abundant (> 40 %)
#> SizeValue ContrastValue BoundaryClassificationValue
#> 1 Fine (2 - 6 mm) Distinct Clear (0,5 - 2 mm)
#> 2 Coarse (> 20) Prominent Clear (0,5 - 2 mm)
#> 3 Fine (2 - 6 mm) Distinct Sharp (< 0,5 mm)
#> 4 Coarse (> 20) Distinct Sharp (< 0,5 mm)
#> 5 Fine (2 - 6 mm) Faint Sharp (< 0,5 mm)
#> 6 Medium (6 - 20 mm) Prominent Clear (0,5 - 2 mm)
#> CarbonatesContentValue CarbonatesFormsValue
#> 1 Slightly calcareous Hard concretions
#> 2 Non-calcareous Disperse powdery lime
#> 3 Strongly calcareous Hard concretions
#> 4 Slightly calcareous Hard cemented layer or layers of carbonates
#> 5 Moderately calcareous Hard concretions
#> 6 Moderately calcareous Disperse powdery lime
#> GypsumContentValue GypsumFormsValue SaltContentValue
#> 1 Moderately gypsiric Soft concretions ST
#> 2 Moderately gypsiric Hard cemented layer or layers of gypsum EX
#> 3 Moderately gypsiric Disperse powdery gypsum MO
#> 4 Moderately gypsiric “gazha” ST
#> 5 Moderately gypsiric Hard cemented layer or layers of gypsum MO
#> 6 Strongly gypsiric Disperse powdery gypsum VST
#> WaterContentSat FieldPh SoilOdourValue
#> 1 MS pHCaCl2 of < 3.6, if > 15% OM None
#> 2 SCL pHCaCl2 of < 3.2, if < 4% OM None
#> 3 SC pHCaCl2 of < 3.4, if 4–15% OM Petrochemical
#> 4 Gravel, CS pHCaCl2 of < 3.6, if > 15% OM None
#> 5 Si pHCaCl2 of < 3.4, if 4–15% OM Sulphurous
#> 6 HC > 60% clay pHCaCl2 of < 3.4, if 4–15% OM Petrochemical
#> AndicCharacteristics OrganicMatterEstimation StructureGradeValue
#> 1 - Grey Munsell value 5,5 Weak to moderate
#> 2 + Dark grey Munsell value 4 Strong
#> 3 + Dark grey Munsell value 4 Strong
#> 4 - Black Munsell value 2 Strong
#> 5 - Light grey Munsell value 6,5 Moderate to strong
#> 6 + Dark grey Munsell value 4 Moderate
#> StructureTypeValue StructureCodeValue StructureSizeValue
#> 1 Platy Lumpy Very fine / thin
#> 2 Rock structure Angular blocky (parallelepiped) Medium
#> 3 Crumbs, lumps and clods Columnar Fine / thin
#> 4 Prismatic Rock structure Fine / thin
#> 5 Rock structure Angular and subangular blocky Very fine / thin
#> 6 Platy Subangular prismatic Medium
#> CombiSoilStructureSize CombiSoilStructure
#> 1 Fine and medium Primary breaking to secondary structure
#> 2 Very fine and fine One structure merging into the other
#> 3 Medium to very coarse Both structures present
#> 4 Medium and coarse One structure merging into the other
#> 5 Very fine and fine Primary breaking to secondary structure
#> 6 Very fine and fine Primary breaking to secondary structure
#> ConsistenceDryValue ConsistenceMoistValue StickinessValue
#> 1 Hard Loose Very sticky
#> 2 Loose friable to firm Non-sticky
#> 3 hard to very hard Extremely firm Non-sticky
#> 4 hard to very hard Firm Non-sticky
#> 5 Slightly hard very friable to friable Non-sticky
#> 6 Very hard Very firm sticky to very sticky
#> PlasticityValue MoistureValue
#> 1 Non-plastic Moist
#> 2 plastic to very plastic Very wet
#> 3 Very plastic Moist
#> 4 Very plastic Very wet
#> 5 Plastic Slightly moist
#> 6 Plastic Very dry
#> BulkDensityMineralValue
#> 1 coherent (prismatic, columnar, wedgeshaped)
#> 2 coherent, prismatic, platy, (columnar, angular blocky, platy, wedgeshaped)
#> 3 prismatic, platy, (angular blocky)
#> 4 coherent, prismatic, platy, (columnar, angular blocky, platy, wedgeshaped)
#> 5 single grain, subangular, angular blocky
#> 6 angular blocky
#> BulkDensityPeatValue PeatDrainageValue PorosityValue VoidsClassificationValue
#> 1 > 0.17 Rather loose Low Vesicular
#> 2 0.11–0.17 Dense Very low Vesicular
#> 3 > 0.17 Dense Very high Channels
#> 4 0.11–0.17 Rather dense Very high Vesicular
#> 5 < 0.04 Loose Low Planes
#> 6 0.11–0.17 Rather loose Very high Vesicular
#> VoidsDiameterValue PoresAbundanceValue CoatingAbundanceValue
#> 1 Fine Very few Few
#> 2 Medium Many Few
#> 3 Very coarse None Few
#> 4 Medium Very few Few
#> 5 Very coarse None Common
#> 6 Very fine Common Abundant
#> CoatingContrastValue CoatingNatureValue CoatingFormValue
#> 1 Faint Clay and sesquioxides Discontinuous irregular
#> 2 Distinct Clay and sesquioxides Discontinuous circular
#> 3 Distinct Clay Discontinuous circular
#> 4 Distinct Jarosite Dendroidal
#> 5 Distinct Clay and sesquioxides Other
#> 6 Faint Manganese Dendroidal
#> CoatingLocationValue CementationContinuityValue CementationFabricValue
#> 1 Coarse fragments Broken Platy
#> 2 Horizontal pedfaces Discontinuous Pisolithic
#> 3 No specific location Continuous Vesicular
#> 4 No specific location Broken Vesicular
#> 5 Lamellae (clay bands) Continuous Vesicular
#> 6 Coarse fragments Broken Vesicular
#> CementationNatureValue CementationDegreeValue
#> 1 Not known Non-cemented and non-compacted
#> 2 Carbonates Non-cemented and non-compacted
#> 3 Ice Non-cemented and non-compacted
#> 4 Clay Compacted but non-cemented
#> 5 Iron-manganese (sesquioxides) Non-cemented and non-compacted
#> 6 Ice Moderately cemented
#> ConcentrationVolumeValue ConcentrationKindValue ConcentrationSizeValue
#> 1 Abundant Other Medium
#> 2 Few Nodule Coarse
#> 3 None Nodule Fine
#> 4 Very few Nodule Fine
#> 5 None Crack infillings Fine
#> 6 Dominant Crystal Very fine
#> ConcentrationShapeValue ArtefactsHardnessValue ConcentrationNatureValue
#> 1 Irregular Soft Carbonates-silica
#> 2 Irregular Both hard and soft Silica (siliceous)
#> 3 Flat Both hard and soft Clay-sesquioxides
#> 4 Angular Soft Not known
#> 5 Irregular Both hard and soft Not known
#> 6 Irregular Both hard and soft Clay-sesquioxides
#> ArtefactsColourValue RootsDiameterValue RootsAbundanceValue
#> 1 Reddish yellow Medium Few
#> 2 Yellowish brown Very fine Few
#> 3 Reddish brown Medium Common
#> 4 Brownish Fine None
#> 5 White Fine Few
#> 6 Reddish brown Very fine and fine None
#> BiologicalAbundanceValue BiologicalFeaturesValue ArtefactsKind
#> 1 Few Infilled large burrows Pavements and paving stones
#> 2 None Charcoal Organic garbage
#> 3 Few Earthworm channels Organic garbage
#> 4 Common Earthworm channels Organic garbage
#> 5 Common Pedotubules Synthetic liquid
#> 6 None Artefacts Organic garbage
#> DepositsValue1
#> 1 Stratified (spoiled materials)
#> 2 Not stratified but clods of different colour, texture and/or artefacts (dumped substrate)
#> 3 Not stratified but clods of different colour, texture and/or artefacts (dumped substrate)
#> 4 Not stratified but clods of different colour, texture and/or artefacts (dumped substrate)
#> 5 Not stratified but clods of different colour, texture and/or artefacts (dumped substrate)
#> 6 Not stratified but clods of different colour, texture and/or artefacts (dumped substrate)
#> DepositsValue2
#> 1 Earthy, humic (grey to blackish grey) --> topsoil material --> ...UA1
#> 2 Mainly sandy --> sand --> ...UU3
#> 3 Clayey --> clay --> ...UU1
#> 4 Clayey --> clay --> ...UU1
#> 5 Mainly broken rock --> broken rock --> ...UU5
#> 6 Dark grey to black, NH3 smell, artefacts --> sewage sludge --> ...UA2
#> soildepthclass siteid
#> 1 major than observed depth siteid_1
#> 2 major than observed depth siteid_2
#> 3 major than observed depth siteid_3
#> 4 major than observed depth siteid_4
#> 5 equal to observed depth siteid_5
#> 6 equal to observed depth siteid_6Simulating genomic module
declaring global information
We need to simulate the I genotypes expression of n markers. We assume that the genetic constitution of the genotypes does not differ across environments.
#our problem is to simulate the expression of 100 traits for each genotype inside each J environments.
known_neg_dis_gen=calculate_trig_dis(c(-0.8016201, -0.8022176, -0.7633010,0.80))
known_pos_dis_gen=calculate_trig_dis(c(0.5,0.6,0.9,0.1))
# we redefine the structure
n_genotypes=10
n_markers= ncol(gen_att)
gen_i_expr=vector("list", length = n_markers)
## we need to insert the levels of genotypes to be considered
genotypes_id=sprintf("genotype_%d",seq(1:n_genotypes))
gen_att=add_to_df(gen_att,genotypes_id)
names(gen_att)[101]="genotype_id"
gen_i_coords=module_coords(n_var=ncol(gen_att),
which_neg = c("1_2","4_5","2_3","20_4","30_3"),
dis_neg_know_ass =known_neg_dis_gen,
coo_known_neg_ass=c("4_5","2_3","20_4","30_3"),
dis_pos_know_ass =known_pos_dis_gen,
coo_known_pos_ass = c("3_9","4_5","6_8","7_8"),
by=0.0001)
## creating the symetric matrices of distances and corrs
sym_tri_gen_i=sym_mtx(vector=gen_i_coords$coords$distance,
n_var=ncol(gen_att),
var_names = colnames(gen_att)
)
sym_sin_gen_i=sym_mtx(vector=gen_i_coords$coords$sin,
n_var=ncol(gen_att),
var_names = colnames(gen_att)
)
gen_i_corr=assure_corr(n_var=ncol(gen_att),corr=sym_sin_gen_i)
gen_i_corr=as.matrix(gen_i_corr)
is.positive.definite(gen_i_corr)
#> [1] TRUE
sim_data_cop_gen_i=set_sim_copula(d=ncol(gen_att),
lower_tri_corr = gen_i_corr[lower.tri(gen_i_corr,diag=FALSE)],
n_cont=0,
cont_var_par = list(),
n_unique=length(unique(na.omit(gen_att$genotype))),
mar_cont_dists=c(),
var_names=colnames(gen_att))
cut_data_gen_i=cut_levels(categorical=gen_att,
cat_inc_all_levels=c("genotype_id"),
levels_inc_all=list(genotype_id=c(1:10)), continuous_to_cut=sim_data_cop_gen_i$simulated,corrs=gen_i_corr
)
## this data contains the expression of the markers
## for each of n_genotypes
head(cut_data_gen_i[1:3,c(1,4,101)])
#> trait_1 trait_4 genotype_id
#> 1 2 1 genotype_id_1
#> 2 1 0 genotype_id_2
#> 3 1 0 genotype_id_3
## the data will repeat across n_sites environments.
n_sites=length(na.omit(soil_att_ex$siteid))
sites_id=sprintf("siteid_%d",seq(1:n_sites))
gens_site_id=rep(sites_id,each=n_genotypes)
abemus_genomics=do.call("rbind", replicate(n_sites, cut_data_gen_i, simplify = FALSE))
abemus_genomics$siteid=gens_site_id
head(abemus_genomics[1:3,c(1,10,101,102)])
#> trait_1 trait_10 genotype_id siteid
#> 1 2 0 genotype_id_1 siteid_1
#> 2 1 1 genotype_id_2 siteid_1
#> 3 1 0 genotype_id_3 siteid_1Merging simulated and genomic simulated data.
#head(cut_data_soil)
#head(abemus_genomics)
final_soil_gen<-merge(abemus_genomics,cut_data_soil, by="siteid")
head(final_soil_gen[c(1,3,120,105),c(1,5,100,102,120,105)])
#> siteid trait_4 trait_99 genotype_id CropClassValue1
#> 1 siteid_1 1 2 genotype_id_1 Oilcrops
#> 3 siteid_1 0 1 genotype_id_3 Oilcrops
#> 120 siteid_5 1 2 genotype_id_10 Fodder plants
#> 105 siteid_4 2 2 genotype_id_5 Oilcrops
#> WeatherConditionsValue_present
#> 1 Sleet
#> 3 Sleet
#> 120 Snow
#> 105 SleetSimulating phenomics
target <- sim_target(
X_gene = dat %>% dplyr::select(`trait_1`:`trait_100`),
X_env = dat %>% dplyr::select(`DranaigeClass`:`yes_no_sample`),
method = "lasso", pars = list(lambda = 0.2, sigma = 1,k=3),
marginal_mean = 5.3, marginal_sd = 1.5
)
dat$target<-target$target
qplot(genotype_id, target, data = dat,
geom= "boxplot", fill = genotype_id)+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))+ggtitle("Simulated values of yield (t/ha) with Lasso")+ylab("yield (t/ha)")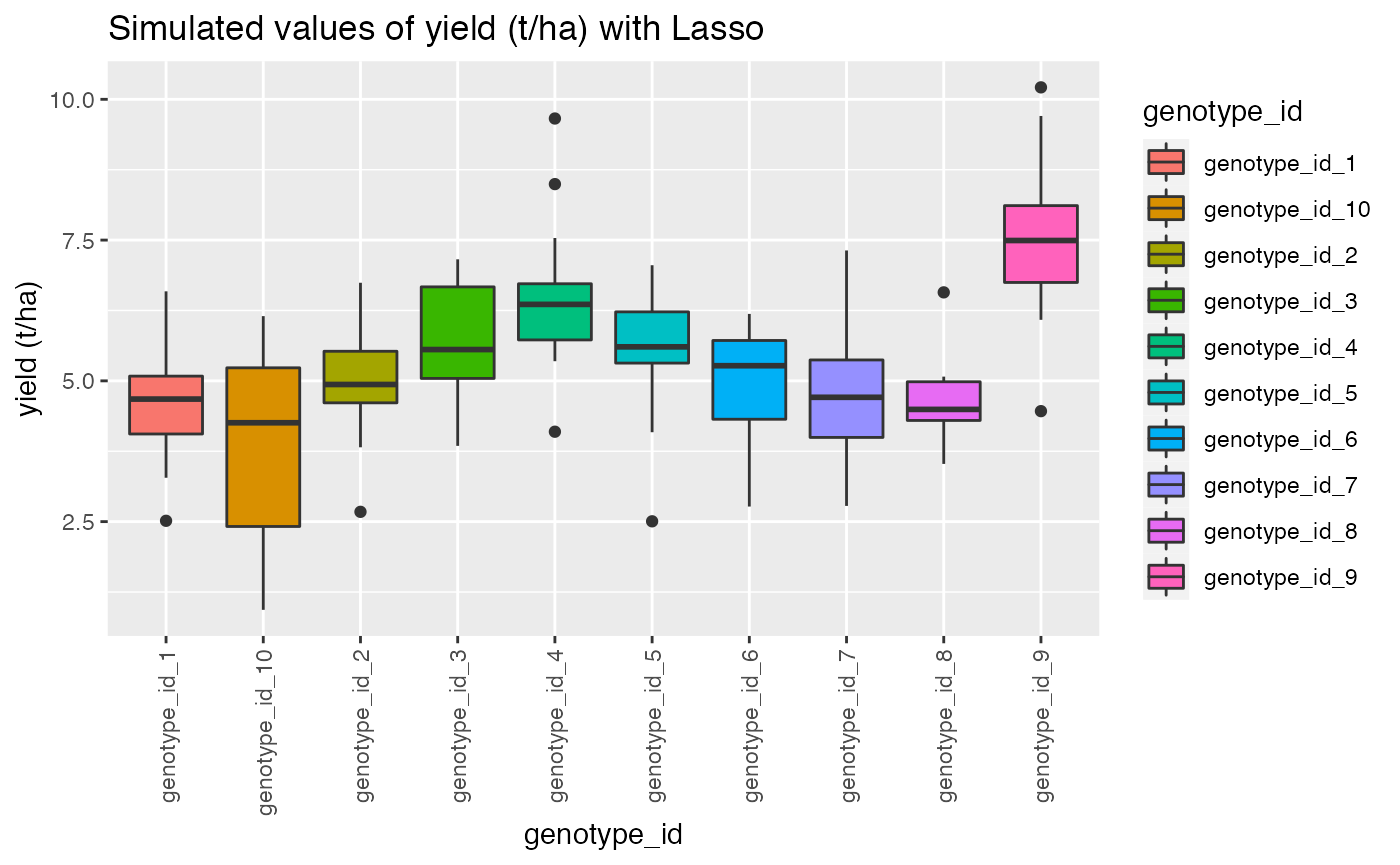
qplot(siteid, target, data = dat,
geom= "boxplot", fill = siteid)+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))+ggtitle("Simulated values of yield (t/ha) with Lasso")+ylab("yield (t/ha)")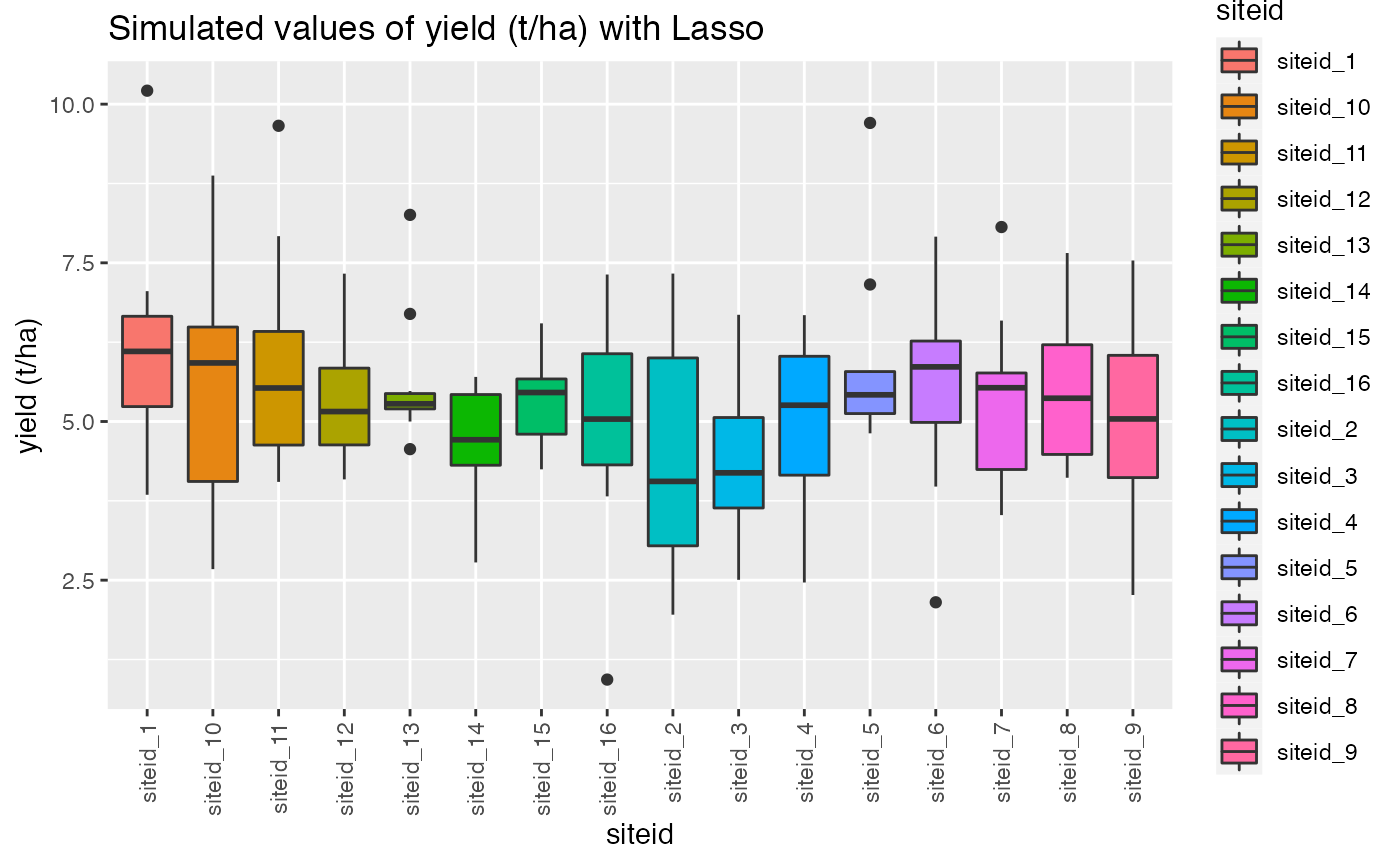
#interaction.plot(dat$genotype, dat$environment, dat$yield, fixed = TRUE)
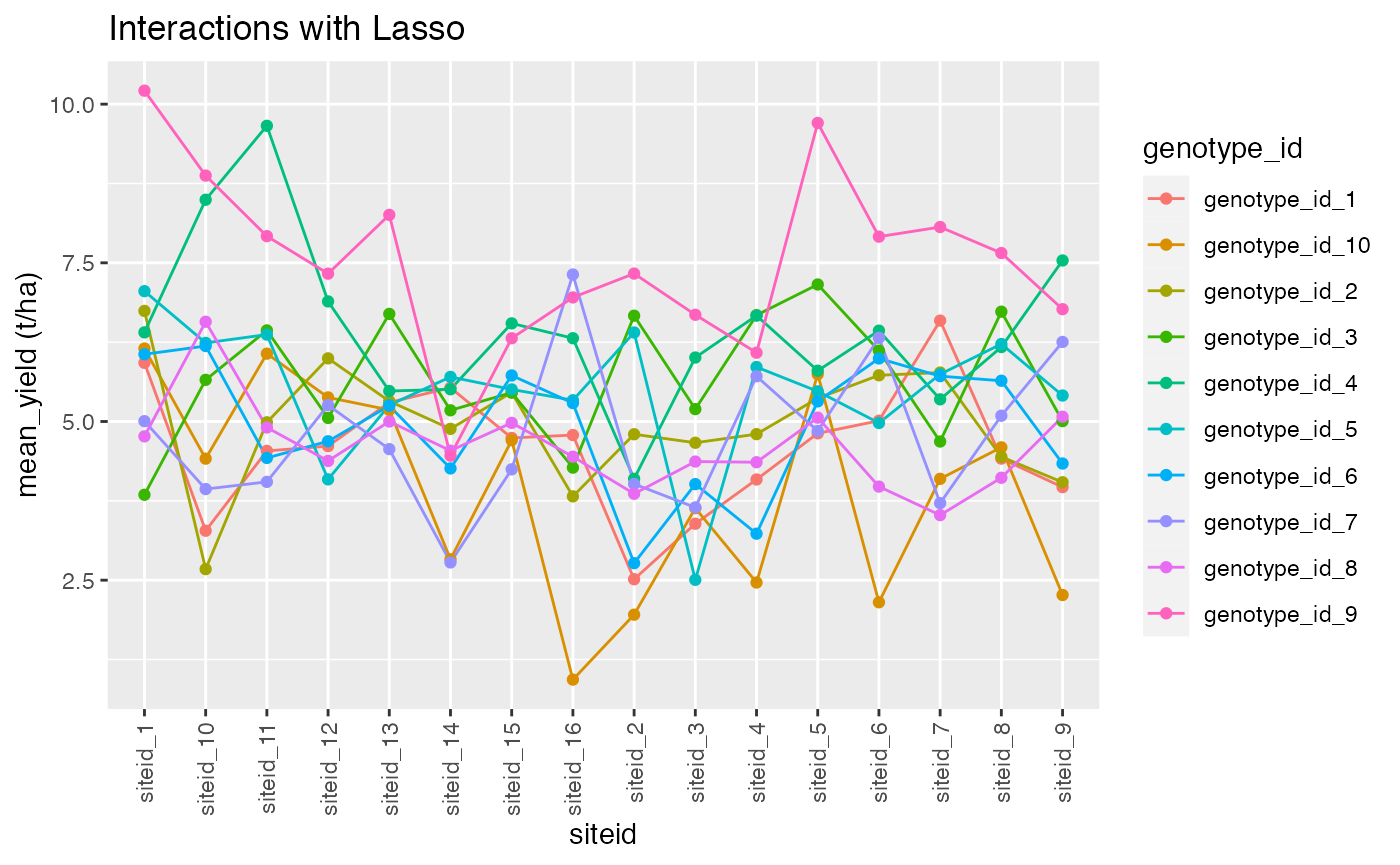
dat %>%
group_by(genotype_id,siteid) %>%
summarise(mean_yield = mean(target)) ->tips2
tips2 %>%
ggplot() +
aes(x = siteid, y = mean_yield, color = genotype_id) +
geom_line(aes(group = genotype_id)) +
geom_point()+ggtitle("Interactions with Lasso")+ylab("mean_yield (t/ha)")+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))